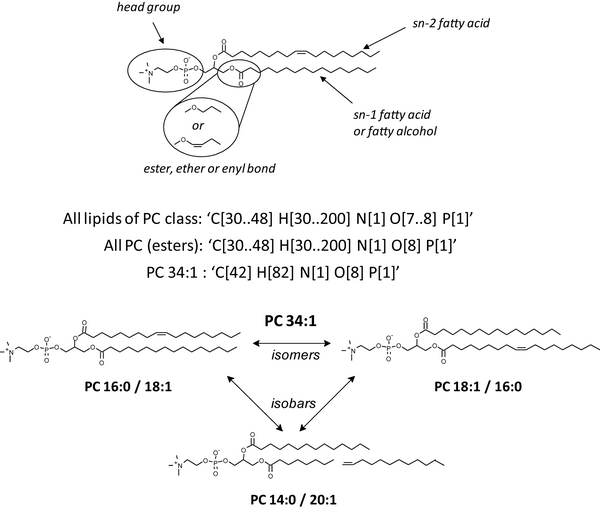
LipidXplorer is a software that supports a variety of untargeted shotgun lipidomics experiments. It uses user-defined MFQL (for Molecular Fragmentation Query Language) rules to identify and quantify lipids. It is designed to support bottom-up and top-down shotgun experiments on all types of tandem MS platforms.
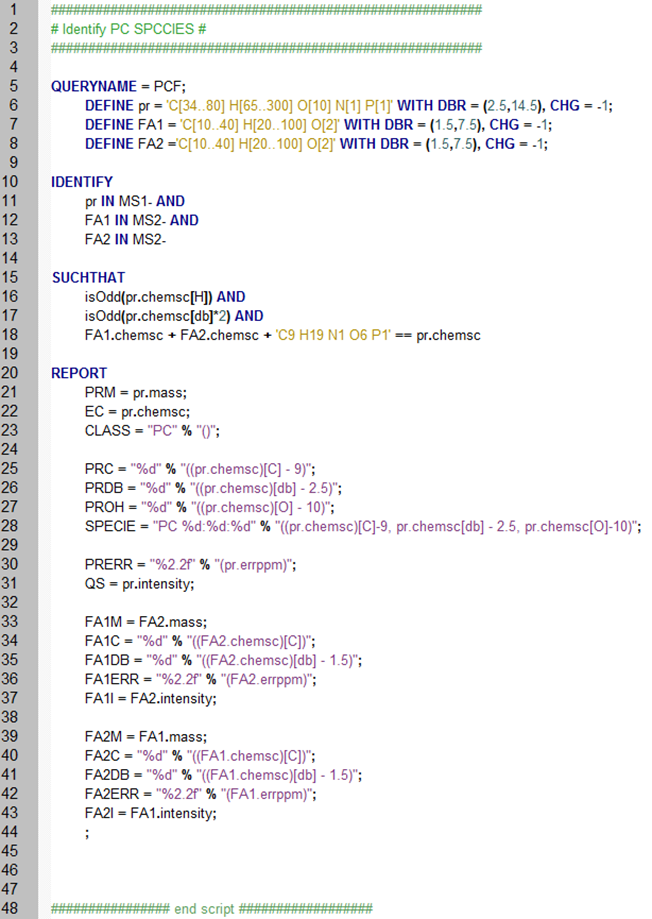
Using MFQL users can customize the complete lipid identification procedure: it starts from DEFINEning elemental compositions of precursor (pr) and fragment (FA1, FA2) ions. Next, the user describes how to IDENTIFY the lipid by looking for precursor and fragments in MS 1 and MS2 spectra, respectively. SUCHTHAT section describes fragments and precursor masses should match. Finally, REPORT section determines how the identified lipids and intensities of corresponding peaks will be made available.
You can find the Desktop version at https://lifs-tools.org/lipidxplorer.html, there you will also have access to the documentation including the wiki (https://lifs-tools.org/lipidxplorer), that can guide you through the initial steps, also if you have any issues you can contact us via https://lifs-tools.org/support.html.
You don’t have to install LipidXplorer to try it, you can use the web version!
LipidXplorer Web: https://apps.lifs-tools.org/lipidxplorer/
There you will find sample files and instruction on how to run LipidXplorer to get an initial introduction.
LipidXplorer2, prepublication access: https://apps.lifs-tools.org/lipidxplorer-dev/
You can also try our new LipidXplorer2, that does not required configuration information, just choose the MFQL and provide the spectra files and LipidXplorer2 does all the rest!
If you are using a particular version of LipidXplorer, from version 1.2.8 onwards, each release now has a citable DOI for LipidXplorer@Zenodo
Please reference LipidXplorer in general by citing the following publications:
Herzog R, Schuhmann K, Schwudke D, Sampaio JL, Bornstein SR, Schroeder M, et al. (2012) LipidXplorer: A Software for Consensual Cross-Platform Lipidomics. PLoS ONE 7(1): e29851. doi:19.1371/journal.pone
Herzog R, Schwudke D, Schuhmann K, Sampaio JL, Bornstein SR, Schroeder M, Shevchenko A (2011) A novel informatics concept for high-throughput shotgun lipidomics based on the molecular fragmentation query language. Genome Biol. 2011;12(1):R8. doi:10.1186/gb-2011-12-1-r8
lipidXplorer2 (pending publication)
The latest LipidXplorer release downloads are available from here.

Contact: mirandaa(at)mpi-cbg.de