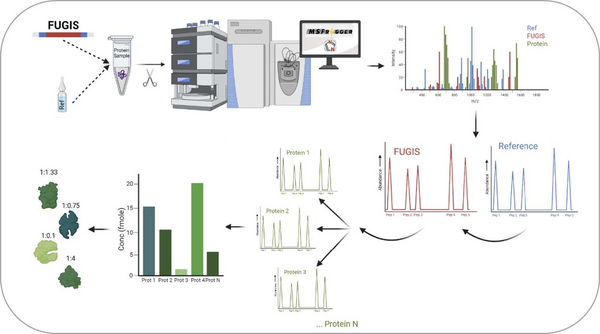
There are accurate methods for molar quantification of proteins, but they are (mostly) targeted – which rely on the likeness of peptide standards to the quantified peptides, and this limits the analytical scope. My goal is to develop a workflow for proteome –wide absolute quantification by DDA (Data Dependent Acquisition) and DIA (Data Independent Acquisition) with a family of FUGIS (Fully Unlabelled Generic Internal Standard) proteins as the internal standard. For the analysis of the complex data, the GlobeQuant software should be adapte and optimized. The optimised workflow will then be applied to understand numerous biomedical problems, such as the molecular composition of lipoproteins, vesicles, organelles and/or entire proteomes.
HongKee Moon (Scientific Computing Facility, MPI-CBG)
