Since 2002 the Myers lab has focused on analyzing and extracting information from images obtained by various forms of microscopy. We believe that such data will reveal more about the function of the entities encoded in the genome then any other approach and will eventually become a prevailing paradigm of investigation, like sequence-based discovery is today. In support of this concept, the group further develops its own customized microscopes.
We have worked on the following representative problems among others:
In broad terms, the computational challenges are to build canonical 3D models of biological systems and map molecular observations onto the model. That is, there is the challenge of registering observations from different animals into a single representative framework, and the challenge of extracting meaningful information in the presence of low SNR and diffraction-limited resolution. The offsetting factor to low SNR and resolution is the presence of very strong prior knowledge about the morphology and dynamics of the entities under observation. Many of our new techniques thus involve what are called template-driven approaches.
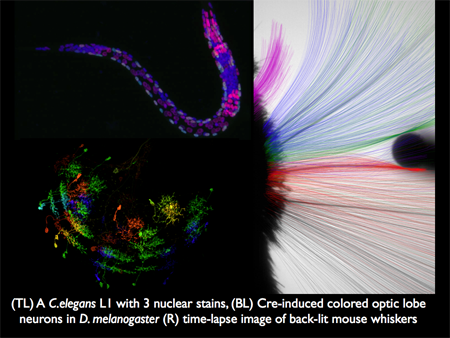
We are in essence a technology group. Our aim is to develop microscopes and software that make observations of in situ and in vivo systems that enable our collaborative partners to advance molecular and cellular biology. We have worked closely with Stuart Kim (Stanford), Chris Doe (Oregon), Tony Hyman (MPI-CBG), Karel Svoboda, Gerry Rubin, Jim Truman, and Tzumin Lee (HHMI JFRC) as examples. The figure below illustrates imagery from three of our projects.
The overarching goal of our group is to build optical devices, collect molecular reagents, and develop analysis software to monitor in as much detail as possible the concentration and localization of proteins, transcripts, and other entities of interest within a developing cohort of cells for the purpose of working together with other groups in the Dresden area towards a biophysical understanding of development at the level of cell communication and force generation.