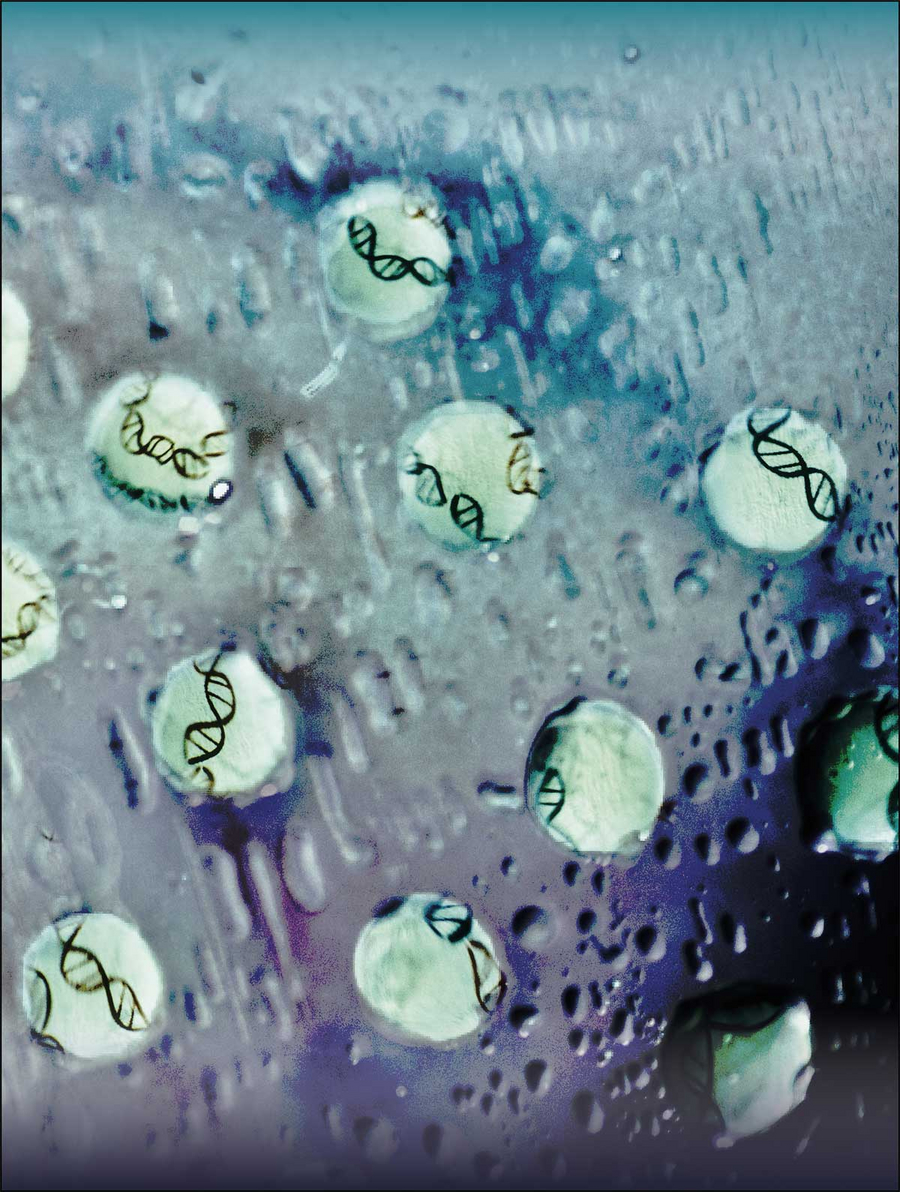
Droplets of transcription factors wet a surface and reveal regulatory DNA regions. This process is symbolized in this photograph of droplets of purified protein on a glass surface with hand-drawn DNA on the back of the glass. Copyright: Mark Leaver, Jose A. Morin and Sina Wittmann, / MPI-CBG / Jose A. Morin et al. Nature Physics, 2022
Life starts with one cell. When an organism develops, dividing cells specialize to form the variety of tissues and organs that build up the adult body, while keeping the same genetic material – contained in our DNA. In a process known as transcription, parts of the DNA – the genes – are copied into a messenger molecule -the ribonucleic acid (RNA) – that carries the information needed to produce proteins, the building blocks of life. The parts of our DNA that are read and transcribed determine the fate of our cells. The readers of the DNA are proteins called transcription factors: they bind to specific sites on the DNA and activate the transcription process. How they recognize which location on the DNA they need to bind to and how these are distinguished from other random binding sites in the genome remains an open question. Scientists at the Max Planck Institute of Molecular Cell Biology and Genetics (MPI-CBG) and the Max Planck Institute for the Physics of Complex Systems (MPI-PKS), both located in Dresden, show that thousands of individual transcription factors team up and interact with each other. They collectively wet the DNA surface by forming liquid droplets that can identify clusters of binding sites on the DNA surface.
Transcription, one of the most fundamental cellular processes, is the action by which the information contained in the DNA is transcribed into the messenger molecule RNA. This "message" is later translated into proteins. Deciding which parts of the DNA are transcribed at any given moment is crucial for proper development to maintain the health of an organism, because many diseases are likely to occur when the genetic programs are not executed correctly. The decision as to which genes are transcribed is made by a complex network of regulatory proteins called transcription factors. While these factors bind to short DNA sequences, the recognition of clusters of many such sequences is required to switch on nearby genes.
The research groups of Stephan Grill and Anthony Hyman, both directors at the MPI-CBG, and the group of Frank Jülicher, director at the MPI-PKS investigated in their recent study in the journal Nature Physics how transcription factors find and recognize clusters of many specific DNA sequences where they can bind and lead to gene activation. To find this out, the researchers followed an interdisciplinary approach, combining expertise in experimental and theoretical biophysics with cell biology. Jose A. Morin, one of the first authors of the study, explains: “We employed optical tweezers – a technology that uses lasers to isolate and manipulate very small objects such as single DNA molecules – combined with confocal microscopy to look at them individually. With optical tweezers it is possible to capture a single DNA molecule and with confocal microscopy we can observe transcription factors binding and forming protein condensates at their preferred DNA sequences. The fact that we can study this process one molecule at the time allowed us to detect interactions otherwise blurred by the complexity of the living cell.” Sina Wittmann, another first author, adds: “With the help of the physicists, we were able to understand how transcription factors communicate with each other and assemble through team work. They undergo what is called a prewetting transition to form liquid-like droplets, which are similar to the drops on a mirror in your bathroom after a shower. These condensates are filled with thousands of transcription factors. Assembled in this way, the transcription factors can now identify the correct DNA region by reading out DNA sequence.”
Stephan Grill summarizes: “We now have a possible mechanistic explanation for the localisation of transcription factors along the genome. This is essential to understand how gene expression is regulated. Since we know that this regulation breaks down in developmental diseases and cancer, these new results give us a clearer picture of how these diseases occur. This knowledge is important to think about new therapeutic options that take the team work of transcription factors into account.”
The research labs of Anthony Hyman, Stephan Grill and Frank Jülicher are also affiliated with both the Center for Systems Biology Dresden (CSBD) and the Cluster of Excellence “Physics of Life” (PoL) at the TU Dresden. The CSBD is a cooperation between the MPI-CBG, the MPI-PKS and the TU Dresden. In the interdisciplinary center, physicists, computer scientists, mathematicians, and biologists work together to understand how cells coordinate their behavior to form tissues and organs of a given form or function. The Cluster of Excellence PoL of TU Dresden seeks to shed light on the laws of physics that underlie the self-organization of life into molecules, cells and tissues. At the cluster, funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation), a cross-disciplinary team of scientists joins forces to investigate how active living matter organizes itself to give rise to life.
Jose A. Morin, Sina Wittmann, Sandeep Choubey, Adam Klosin, Stefan Golfier, Anthony A. Hyman, Frank Jülicher, Stephan W. Grill: Sequence dependent surface condensation of a pioneer transcription factor on DNA. Nature Physics. Doi: 10.1038/s41567-021-01462-2
Media Contact MPI-CBG
Katrin Boes
+49 (0) 351 210 2080
kboes(at)mpi-cbg.de
Further Information:
Prof. Stephan Grill
grill(at)mpi-cbg.de