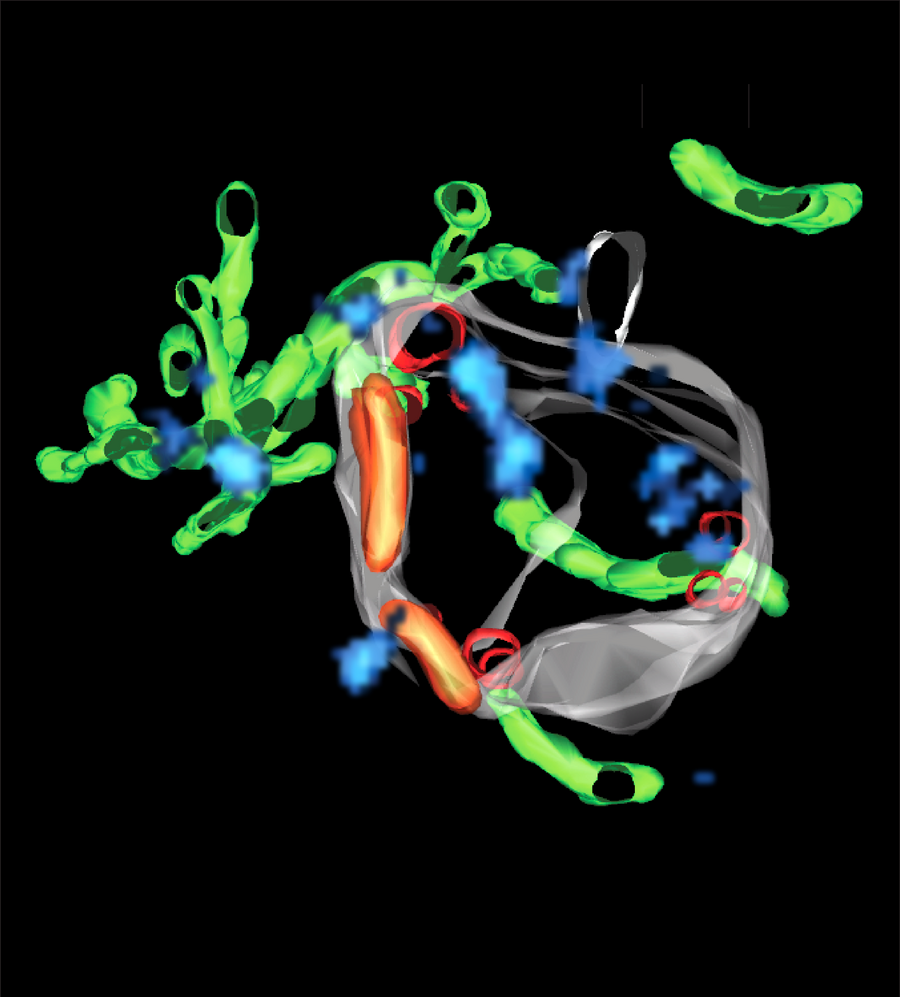
How superCLEM sees an endosome: a triple color SMLM image and the corresponding three-dimensional reconstruction of the entire endosome, based on an electron tomogram, are used to create a detailed model of an early endosome at the nanoscale. It reveals different, structurally distinct endosomal nano-domains, such as small vesicles inside (red) and sorting domains on the outside of the endosomal membrane (orange), delicate tubules attached to the endosomal body(green) and Rab5 nano-domains on the surface of the endosome (blue).
The cell has still a lot of secrets to reveal. The pioneering work with electron microscopy in the 1960s made discoveries on the structure and function of subcellular organelles possible. Since then, scientists have made a lot of progress on the molecular mechanisms whereby organelles are formed and how they ‘talk’ to each other. Cellular organelles are not just bags of molecules but have different shapes. Endosomes for example are membrane-bound organelles and have a shape that resembles an octopus, where the tentacles are tubules to recycle molecules back to the cell surface. The function of endosomes is to receive molecules at the surface of the cell and distribute them within the cell, like a post-office.
So far, it was not possible to create a high precision map of cargo molecules and those that confer specific functions on the endosomes. To do so, new methods are needed that allow visualization of different molecules simultaneously. Several powerful methods exist already. Light microscopy is widely used in cell biology but is limited to 200 nanometres (0.0002 mm) resolution, due to the diffraction of light. Therefore, it cannot resolve the details of many organelles. Another method, single molecule localization microscopy (SMLM), is a super-resolution imaging method that offers an almost molecular resolution down to 20 nanometres (0.00002 mm). However, it cannot capture the entire structure of an organelle. With electron microscopy (EM) on the other hand one can see the whole structure of an organelle but it is difficult to visualize its molecular components.
To overcome this problem, Marino Zerial, director at the MPI-CBG, assembled a team of cell biologists and light and electron microscopists to combine triple colour SMLM with three-dimensional electron tomography in a novel workflow, called superCLEM. To showcase the superCLEM method and workflow, the researchers investigated endosomes, visualizing multiple molecules at the same time without disturbing the delicate fine-structure of these organelles. Christian Franke, Sandra Segeletz and Urska Repnik, three of the authors of the study explain: “So far, it was never possible to simultaneously visualize several specific proteins with nanometer resolution in context with each other on the three-dimensional structure of an organelle.” They continue: “The most important finding is, that the Rab5 protein, which is a key component of endosomes, is not randomly distributed but clustered in small regulatory regions that function like control centers on the endosome membrane. This has never been shown before with such a precision.”
The superCLEM workflow opens up a new field of possibilities to perform structure-function analyses on a wide range of cell organelles at the nanoscale. The method is highly adaptable to many different settings. 3D SMLM methods and alternative EM approaches will broaden superCLEM even further in the future.
Franke, C, Repnik, U, Segeletz, S, et al. Correlative single‐molecule localization microscopy and electron tomography reveals endosome nanoscale domains. Traffic. 2019; 20: 601– 617.