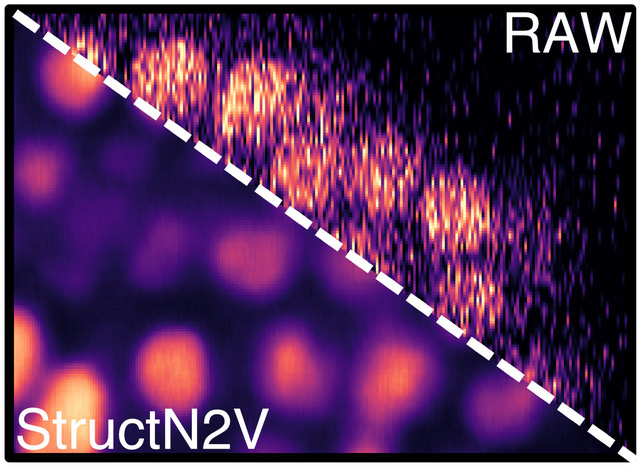
Self-supervised StructN2V removes vertical stripe noise from confocal image of developing C. elegans embryo. © Waterston Lab, University of Washington, Seattle, WA, USA via celltrackingchallenge.net
Fluorescence microscopy has become an indispensable tool to answer fundamental questions in the biomedical sciences. Such microscopes visualize the position of fluorescently labeled cells in biological tissues and can record every single cell in entire developing organisms such as worms, fish, or mice. Just like regular photography, fluorescence microscopy requires a sufficient amount of light. However, the amount of light needed can easily reach levels that harm our model organisms. To avoid that we try to use as little light as possible, which - just like with night photography - greatly amplifies noise and grainy texture in our images. This is a huge problem for biologists trying to see and interpret what the cells are doing.
Progress in using artificial neural networks over the last few years has led to an explosion of image restoration methods for removing noise and improving image quality. Such trained neural networks can “reveal” the content hidden in low-quality microscopy images and restore high-quality microscopy images even if acquired with little light.
Researchers from the Gene Myers group at the MPI-CBG and the neighboring Center for Systems Biology Dresden (CSBD) have now further developed the promising “blind-spot” approach created by their colleagues in the group of Florian Jug (CSBD and MPI-CBG). The new Structured Noise2Void (STRUCTN2V) method by the Myers lab extends the blind-spot approach to remove a wider variety of noise and unwanted texture.
Coleman Broaddus, first author of the study recently published in the conference proceedings of the 2020 IEEE 17th International Symposium on Biomedical Imaging (ISBI), explains: “A blind-spot network is trained by showing it a piece of your image, but hiding a pixel in the middle (the blind spot) and asking the network to guess it’s value. Surprisingly, we found that when the noise has a certain structure carefully hiding extra pixels around the blind spot (thus giving the network even less information to use), actually leads it to do a better job at guessing the right value.”
Gene Myers, director at the CSBD and the MPI-CBG and supervisor of the work, adds: “Our approach fits nicely with the existing denoising approaches already available in the open source library “Noise2Void” maintained by the Jug lab and used by researchers all over the world. We want people to try our approach on their data with as little effort on their part as possible. Hopefully, by making it widely available we will explore and learn about the limits of the technique, while at the same time helping as many people as possible to improve the quality of their microscopy images.”
Coleman Broaddus, Alexander Krull, Martin Weigert, Uwe Schmidt, Gene Myers: „Removing Structured Noise With Self-Supervised Blind-Spot Networks“, Proceedings of the IEEE 17th International Symposium on Biomedical Imaging (ISBI). 2020. DOI: 10.1109/ISBI45749.2020.9098336
Link to open source library: https://github.com/mpicbg-csbd/structured_N2V_paper/