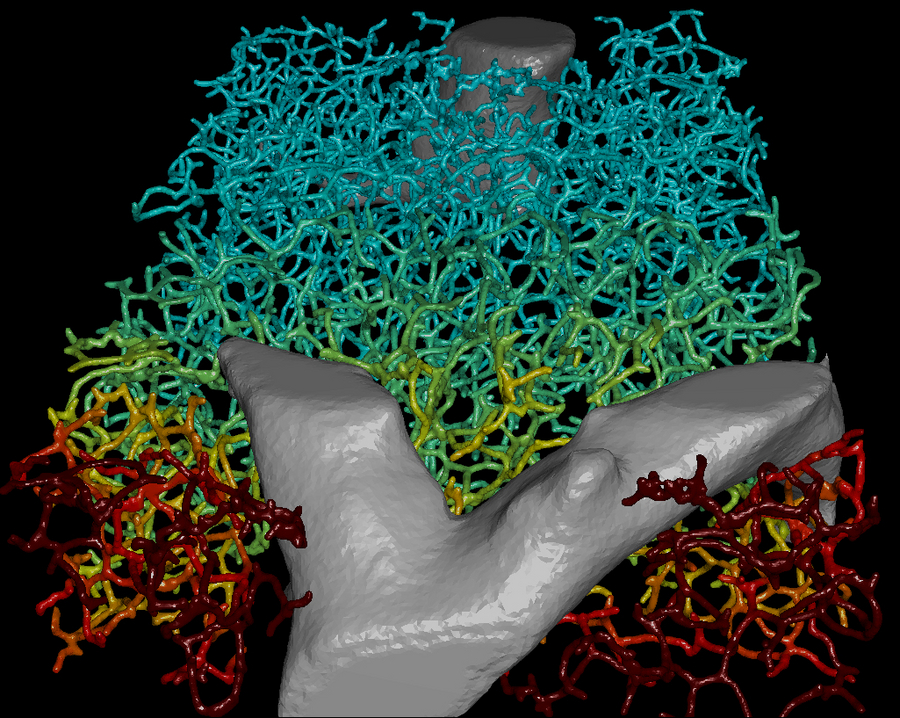
A 3D representation of the geometry of the biliary network with the superimposed bile velocity profile shown as color-code (blue: low velocity; red: high velocity). Picture credits: MPI-CBG
Researchers of the Max Planck Institute of Molecular Cell Biology and Genetics (MPI-CBG) in Dresden developed a multi-scale model that can simulate the fluid dynamic properties of bile in the liver. This model can help to characterize liver diseases as well as drug-induced liver injury and, therefore, is a promising tool for drug development to test and predict the effects of pharmacological compounds on the liver. The research team is now working on a strategy to calibrate this model to human biliary fluid dynamics.
The liver is the central metabolic organ and is crucial for the detoxification of our body. In order to digest lipids and excrete waste products, bile is produced in the liver and transported through a highly ramified tubular network to the intestine. A research team headed by Prof. Marino Zerial, Director at the MPI-CBG, combined high-resolution imaging of the bile canalicular network in the mouse liver with multi-resolution 3D analysis of its geometry to develop a predictive 3D multi-scale model that can simulate the fluid dynamics of bile on several levels – from the sub-cellular to the tissue level. This provides a tool to analyze and understand cholestatic liver disease and to test the effect of pharmacological compounds on bile transport.
So far, the model has only been applied to the mouse. But Zerial and colleagues expect it to be translatable to the human liver: „It is going to be technically challenging, but we now work on a strategy to calibrate our model to human conditions.” This will enable a better understanding of liver diseases and allow to test the effects of pharmacological compounds on the liver. „We demonstrated the precision of our model using Paracetamol as an example: The simulation accurately predicted the effects of a sub-toxic dose of Paracetamol on bile transport.” The model is a promising tool for pharmacological drug development – potential side effects of drugs on bile transport could be predicted and tested more precisely.
The project is a cooperation with the Centre for Information Services and High Performance Computing at Dresden University of Technology as well as with the National Institute of Health. The results of the study were published in „Cell Systems“ and are featured on the cover of the magazine.
Kirstin Meyer, Oleksandr Ostrenko, Georgios Bourantas, Hernan Morales-Navarrete, Natalie Porat-Shliom, Fabian Segovia-Miranda, Hidenori Nonaka, Ali Ghaemi, Jean-Marc Verbavatz, Lutz Brusch, Ivo F. Sbalzarini, Yannis Kalaidzidis, Roberto Weigert, Marino Zerial:
A Predictive 3D Multi-Scale Model of Biliary Fluid Dynamics in the Liver Lobule
Cell Systems, 22 March 2017 (published online 16 March 2017)
doi: 10.1016/j.cels.2017.02.008