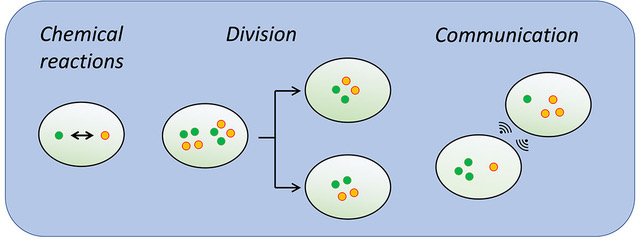
Describing several processes in a biological system with the novel framework. Above, a population of cells evolves as a result of chemical reactions inside them (left), cell-division events (center) and interactions with other cells (right). Copyright: Duso / CSBD / MPI-CBG
Living beings organize their biochemical programs within compartments, such as cells or organelles. However, unlike in a test tube, these reaction containers change with time, for instance when cells grow or divide, or when two organelles inside a cell fuse with one another. The complex interplay between chemical reactions and compartment dynamics governs the behavior of many biological systems, such as a growing cell population, or a network of vesicles that collectively process their molecular cargo.
However, understanding compartmentalized reaction systems through mathematical models is very demanding and effective computational approaches have been lacking so far.
Researchers from the group of Christoph Zechner, group leader at the Center for Systems Biology and the MPI-CBG, have now developed a computational framework, which addresses these difficulties. Their approach, recently published in PNAS, permits to analyze the statistical properties of compartmentalized reaction systems in a very effective manner.
Lorenzo Duso, PhD student in Christoph’s group, and first author of the paper explains: “The main challenge with compartmentalized reaction systems is that they involve an enormous number of variables, which render their numerical analysis very time-consuming. In contrast, our method accumulates these variables into a few summary statistics, which provide a much lower-dimensional description of the system. Thus, we can analyze it by solving just a couple of equations.” The physicist adds: “In the case of vesicle trafficking, for instance, individual vesicles can be created, fuse with each other or split apart, while processing their molecular cargo; it is like an automated warehouse where cargo is steadily re-distributed across many moving boxes. Our method combines all these events and predicts how many compartments we will have in the future and how many molecules of a certain type they will carry.”
Christoph Zechner continues: “Compartmentalization is an important feature of many biological systems and our method helps to make such systems computationally accessible. We thus anticipate many potential applications to biological systems at the subcellular and cellular scale.”
Lorenzo Duso and Christoph Zechner: “Stochastic reaction networks in dynamic compartment populations“, PNAS, 31 August, 2020