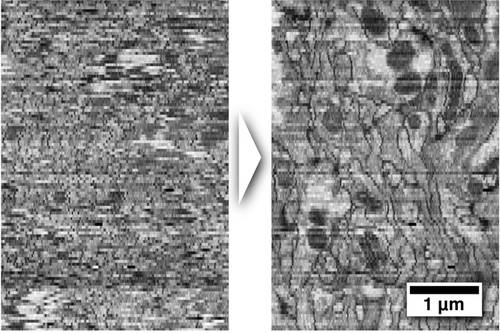
Cross-section through an Electron Microscopy section series of a Drosophila larval brain before (left) and after (right) deformation has been removed.
Anatomy of large biological specimens is often reconstructed from serially sectioned volumes imaged by high-resolution microscopy. It is unavoidable that the sections experience independent deformation during this process, particularly when they are extremely thin (down to 40nm). These deformations make it a difficult challenge to reconstruct 3D anatomy at high precision. MPI-CBG researchers have developed a method to computationally remove these deformations from the sections such that the original volume can be restored.
The method is publically available through the Open-Source image processing platform Fiji. Its main application is the reconstruction of the microcircuitry of biological neuronal tissue from serial section Transmission Electron Microscopy and Array Tomography.
The project is a collaboration with Albert Cardona, formerly at the Institute of Neuroinformatics in Zurich, now at HHMI Janelia Farm. The Transmission Electron Microscopy series was generated by Richard Fetter at HHMI Janelia Farm, the Array Tomography series comes from Stephen Smith's lab at Stanford.
The article is published in Nature Methods.
Stephan Saalfeld, Richard Fetter, Albert Cardona & Pavel Tomancak:
Elastic volume reconstruction from series of ultra-thin microscopy sections
Nature Methods, 12 June 2012