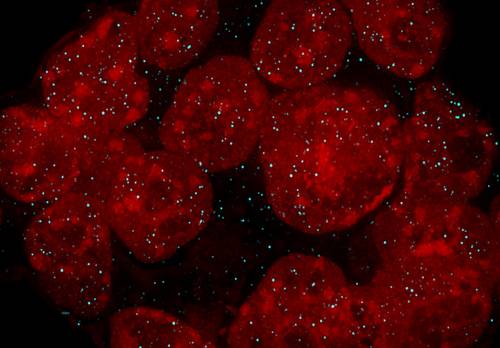
Mouse embryonic stem cells with nuclei in red and long non coding RNA marked in cyan.
RNA Interference (RNAi) has become a standard technique to study gene function and is also a promising approach to find new therapies for a number of diseases. However, so far the RNAi method has only been applied in order to knock down protein-coding genes; techniques to systematically investigate long noncoding RNAs, which have been implicated in diverse biological pathways, are limited.
A team of researchers led by Frank Buchholz have now developed a method to design and produce RNAi and localization resources for long coding RNAs, enabling functional characterization to study these molecules. Long non-coding RNAs are of particular interest because they are known to participate in various biological processes such as maintenance of embryonic stem cell (ESC) identity or the regulation of disease states.
Debojyoti Chakraborty, Dennis Kappei, Mirko Theis, Anja Nitzsche, Li Ding, Maciej Paszkowski-Rogacz, Vineeth Surendranath, Nicolas Berger, Herbert Schulz, Kathrin Saar, Norbert Hubner & Frank Buchholz:
Combined RNAi and localization for functionally dissecting long noncoding RNAs
Nature Methods (2012), doi:10.1038/nmeth.1894, 12 February 2012