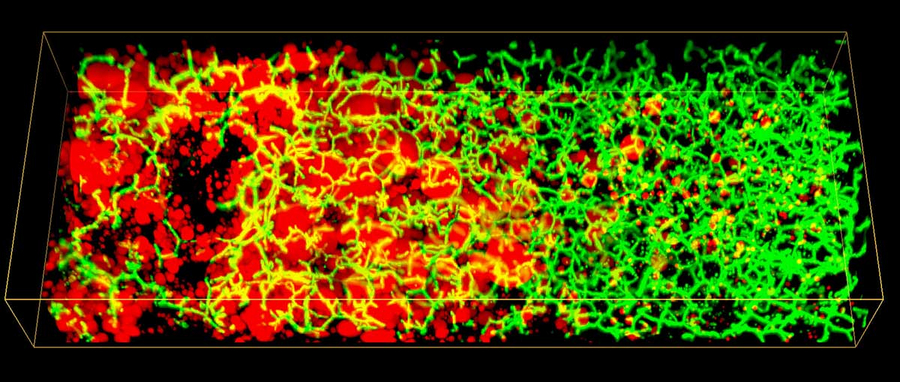
3D image of lipid droplets (red) and Bile Canaliculi network (green) along the liver lobule of a patient with non-alcoholic fatty liver disease (NAFLD). Image credits: Hernán Morales-Navarrete. Copyright: Segovia-Miranda et al. / MPI-CBG
Non-alcoholic fatty liver disease (NAFLD) is becoming the most common chronic liver disorder in developed countries. Histological analysis of liver tissue is the only widely accepted test for diagnosing and distinguishing different stages of NAFLD. However, this technique provides only two-dimensional (2D) images of the liver tissue in low resolution and overlooks potentially important 3D structural changes. Researchers at the Max Planck Institute of Molecular Cell Biology and Genetics in Dresden (MPI-CBG) and the University Hospital Carl Gustav Carus Dresden (UKD) together with colleagues from the Technische Universität Dresden (TUD) now generated 3D geometrical and functional models of human liver tissue for different stages of NAFLD which reveal new critical tissue alterations, providing new insights into NAFLD pathophysiology and contributing to high definition medical diagnosis. The results are published in the journal Nature Medicine.
Non-alcoholic fatty liver disease (NAFLD) is characterized by the accumulation of fat in the liver with an insulin resistance due to causes other than alcohol intake. NAFLD includes a spectrum of liver diseases from simple steatosis (“non-progressive” and reversible) to non-alcoholic steatohepatitis (NASH), which can progress into cirrhosis, liver cancer, or liver failure, requiring eventually transplantation. In 2017, an estimated 24% of the worldwide population was affected by NAFLD, which makes it the leading cause of chronic liver disease. Conventional histological analysis of liver tissue is the gold standard for diagnosing NAFLD progression, but it has several disadvantages: The low resolution 2D images of liver tissue only allow a semi-quantitative evaluation and it can be subjective, since it depends on the pathologist’s skills. Most importantly, it fails to provide 3D information on tissue structure and function.
In order to overcome those limitations in diagnosis, the research team of MPI-CBG director Marino Zerial together with colleagues from the Department of Medicine I, Gastroenterology and Hepatology at the UKD, from the University hospitals in Rostock and Kiel, and from the Center for Information Services and High Performance Computing (ZIH) at the TUD developed 3D spatially resolved geometrical and functional models of human liver tissue at different stages of NAFLD. The liver has a complex 3D tissue organization: It consists of functional units, the liver lobuli, containing two intertwined networks, the sinusoids for blood flow and the bile canaliculi for bile secretion and flux. Such an architecture makes it difficult to grasp the 3D organization and overall tissue structure from 2D histological images.
In 2017, the research group of Marino Zerial developed a model of the bile canalicular network and bile flow in the mouse liver using multi-resolution 3D analysis of its geometry. Now, the team examined the 3D organization of human liver tissue. Although several defects can already be seen in 2D images, the alterations of bile canaliculi and sinusoidal networks can only be recognized upon a 3D reconstruction. Fabián Segovia-Miranda, the first author of the study reports: “Recent advances in making tissue transparent and multi-photon microscopy allow for the imaging of thicker tissue sections, so that 3D information can be captured.” The 3D digital reconstruction of those tissues was then used to computationally simulate bile fluid dynamics through a model created by the ZIH. Lutz Brusch from the department of innovative methods of computing at the ZIH explains: “While the fluid dynamics of blood flow through the comparatively large capillaries has been addressed by simulations, this was so far impossible for bile due to the lack of accurate geometrical data of human tissue across all relevant scales.” With microscopy, digital image reconstruction and computational modelling combined, the researchers identified a set of cellular and tissue parameters connected with NALFD progression. Fabián adds: “We discovered that the structure of the 3D bile canaliculi network is profoundly different in NAFLD tissue. Such structural changes have also critical functional consequences. Using personalized biliary fluid dynamic simulations, we learned that the flow of bile in some small areas of the tissue is compromised, which is called micro-cholestasis.”
Marino Zerial, who is also affiliated with the Center for Systems Biology Dresden (CSBD), gives an outlook: “High definition medicine paves the way towards diagnosing diseases such as NAFLD at early stages, much before symptoms appear. It also helps us to identify molecular pathogenetic mechanisms to design novel therapies.” Jochen Hampe, professor at the UKD, adds: “This three-dimensional analysis of liver tissue allows us to gain completely new insights into the disease mechanisms. This enables us to better understand how bile flow and disease progression are related. It also opens up new approaches for therapies.”
Fabián Segovia-Miranda, Hernán Morales-Navarrete, Michael Kücken, Vincent Moser, Sarah Seifert, Urska Repnik, Fabian Rost, Mario Brosch, Alexander Hendricks, Sebastian Hinz, Christoph Röcken, Dieter Lütjohann, Yannis Kalaidzidis, Clemens Schafmayer, Lutz Brusch, Jochen Hampeand Marino Zerial: “3D spatially-resolved geometrical and functional models of human liver tissue reveal new aspects of NAFLD progression.” Nature Medicine, 02. December 2019. Doi: 10.1038/s41591-019-0660-7