Biology is well equipped in exploiting a large number of out of equilibrium processes to support life. A complete understanding of these mechanisms is still in its infancy due to the complexity and number of the individual components involved in the reactions. A bottom up approach allows us to replicate key biological processes using a small number of basic building blocks. This methodology has the added advantage that properties and characteristics of the artificial cell can be readily tuned and adapted. Working between biophysics, materials science and synthetic biology we re-imagine and translate the physical phenomena which drive out of equilibrium processes in cells into novel, robust and dynamic systems to address central questions in the Biological Sciences and the Origin of life. “ How do biological cells and tissues sustain life from collections of non-living molecules?”. We are active within the synthetic cell community and aim to exploit our minimal systems for synthetic biology applications.
We aim to build synthetic cellular systems which can sustain out-of-equilibrium behaviour. We are inspired by biological cells and aim to incorporate features of compartmentalisation, reactions and communication into our synthetic cellular systems.
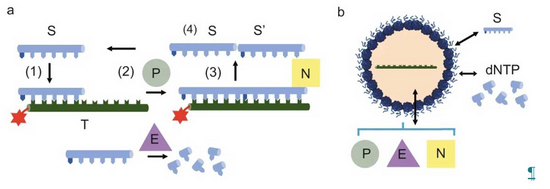
A key property of the cell is, it’s ability to compartmentalize chemical reactions. This allows the cell to control different chemical and physical environments. Membrane bound and membrane free compartmentalisation play a role in biological systems to delineate distinct biochemical environments.
In order to understand how compartmentalisation can tune and regulate biochemical reactions, we have compiled a synthetic cellular toolkit. This toolkit contains membrane bound compartments based on the self-assembly of lipids (vesicles) or proteinosomes (protein-polymer conjugates) and membrane free compartments formed via liquid-liquid phase separation processes (coacervation). Our toolkit also contains a wide variety of enzyme reactions from primitive RNA reactions to cell free transcription and translation and PEN DNA reactions. With our toolkit we use quantitative approaches combined with physical modelling to unravel the physico-chemical principles by which enzyme reactions can be tuned by compartmentalisation.
Selected publications:
Björn Drobot, Juan M Iglesias-Artola, Kristian Le Vay, Viktoria Mayr, Mrityunjoy Kar, Moritz Kreysing#, Hannes Mutschler#, T-Y Dora Tang#. Compartmentalised RNA catalysis in membrane-free coacervate protocells. Nat Commun, 9(1) Art. No. 3643 (2018).
Juan M Iglesias-Artola, Björn Drobot, Mrityunjoy Kar, Anatol Fritsch, Hannes Mutschler, T-Y Dora Tang, Moritz Kreysing. Charge-density reduction promotes ribozyme activity in RNA-peptide coacervates via RNA fluidization and magnesium partitioning. Nat Chem, 14(4) 407-416 (2022).
Adrian Zambrano✳︎, Giorgio Fracasso✳︎, Mengfei Gao✳︎, Martina Ugrinic, Dishi Wang, Dietmar Appelhans, Andrew deMello, T-Y Dora Tang. Programmable synthetic cell networks regulated by tuneable reaction rates. Nat Commun, 13(1) Art. No. 3885 (2022).
Mengfei Gao, Dishi Wang, Michaela Wilsch-Bräuninger, Weihua Leng, Jonathan Schulte, Nina Morgner, Dietmar Appelhans, T-Y Dora Tang. Cell Free Expression in Proteinosomes Prepared from Native Protein-PNIPAAm Conjugates. Macromol Biosci, Art. No. e2300464 (2023).
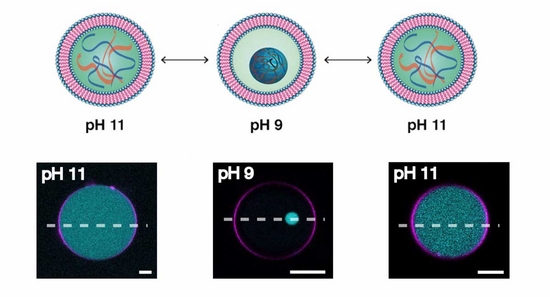
In nature, an out of equilibrium state is maintained by taking energy from the environment and exploiting biochemical network complexity. As minimal synthetic cells lack this complexity, we seek new strategies to achieve and sustain out-of-equilibrium behaviour within synthetic cellular systems. In addition, we explore the role of compartmentalisation on sustaining out-of equilibrium dynamics. In this area, we plan to define the minimal number of components required to sustain dynamic behaviour within synthetic cellular systems.
Selected publications:
Celina Love, Jan Steinkühler, David Thomas Gonzales, Naresh Yandrapalli, Tom Robinson, Rumiana Dimova, T-Y Dora Tang. Reversible pH-Responsive Coacervate Formation in Lipid Vesicles Activates Dormant Enzymatic Reactions. Angew Chem Int Ed Engl, 59(15) 5950-5957 (2020).
Alan Ianeselli, Damla Tetiker, Julian Stein, Alexandra Kühnlein, Christof Mast, Dieter Braun#, T-Y Dora Tang#. Non-equilibrium conditions inside rock pores drive fission, maintenance and selection of coacervate protocells. Nat Chem, 14(1) 32-39 (2022).
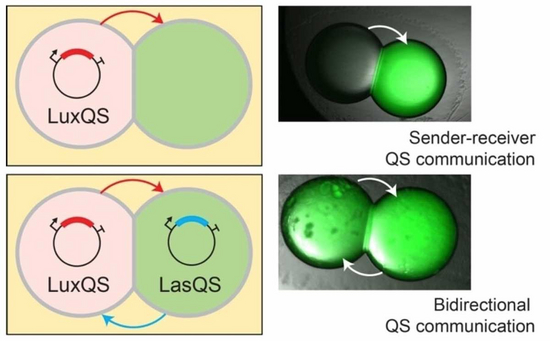
Biological tissues are inherently variable in their cellular compositions. In light of this variability, cells and tissues must co-ordinate their functions to provide robust outcomes to the system. This is an important feature within synthetic cellular systems to ensure robust outcomes despite intrinsic variability. We develop new tools and methods to reduce variability in synthetic cellular systems through engineering approaches. One strategy is to exploit microfluidics to reduce concentration and size variability within synthetic cellular systems. Another strategy relies on exploiting communication by diffusion of molecules to co-ordinate behaviours within populations of synthetic cells.
Selected publications:
T-Y Dora Tang, Dario Cecchi, Giorgio Fracasso, Davide Accardi, Angelique Coutable-Pennarun, Sheref S Mansy, Adam W Perriman, J L Ross Anderson, Stephen Mann. Gene-Mediated Chemical Communication in Synthetic Protocell Communities. ACS Synth Biol, 7(2) 339-346 (2018).
David Thomas Gonzales, Naresh Yandrapalli, Tom Robinson, Christoph Zechner#, T-Y Dora Tang#. Cell-Free Gene Expression Dynamics in Synthetic Cell Populations. ACS Synth Biol, 11(1) 205-215 (2022)
David Thomas Gonzales, Surased Suraritdechachai, Christoph Zechner, T-Y Dora Tang. Bidirectional Communication between Droplet Interface Bilayers Driven by Cell-Free Quorum Sensing Gene Circuits. ChemSystemsChem, 5 Art. No. e20230002 (2023).