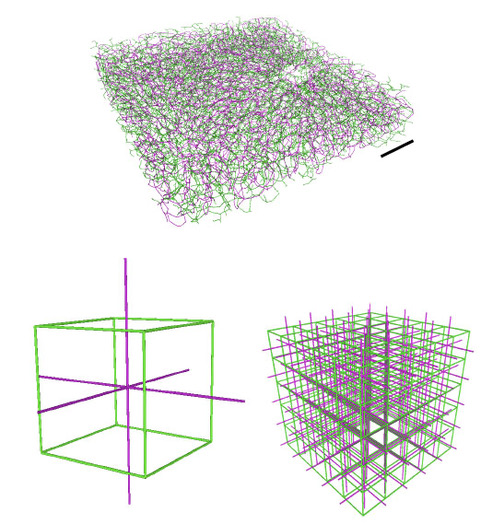
Modeling the growth of intertwined flow networks. TOP: Segment of bile canaliculi (green) and blood vessels (magenta) in the liver (scale bar: 0.2 mm) BOTTOM: Simulation of liver networks as abstracted cubic lattice. Neighbouring edges of each graph enhance or block each others development. © Felix Kramer, CSBD / MPI-CBG
Newly developing blood vessels, e.g. in embryos or during wound healing, are known to first overgrow. Then, dependent on their perfusion, they are cut down and remodeled into a complex network of tiny blood vessels, that ramifies through the tissue. Studies in computational physics have shown that this very process can be simulated with a simple cost model, which describes the balance of overall vessel volume and hydrodynamic resistance. Furthermore, this approach can even predict the relative vessel sizes at branching points, which is known as “Murray’s law.”
Yet, many organs in mammals are not only perfused by the smallest blood vessels (capillaries) possible, but also by even smaller structures, so called secretion networks. One such tiny vessel network, is found in the liver (bile canaliculi) and secrets the bile, which is needed to digest fatty food, but also for cellular waste removal. Though seemingly delicate, these two fluid networks are deeply entangled and mesh like, which enables the liver to be an efficient filter organ. So far, the exact process of generating and maintaining such entangled and mesh like vessel systems, as found in the liver, was still unknown. Researchers from the Carl Modes group at the Center for Systems Biology Dresden (CSBD) and the MPI-CBG have now developed an extended cost model, which couples two flow networks at once. Their study was recently published in Physical Review Research.
Felix Kramer, first author of the paper explains: “Intertwined vessel systems influence each other’s growth, they either block or enhance each other. Our extended model is able to predict the transition from tree-like to mesh graphs, in dependence of the respective interaction of the two networks.” The 3rd year PhD student adds: “Murray’s law works well for single tree-like structures, our approach also allows to describe and quantify the meshes, which wasn’t possible before.”
The altered form of Murray’s law also allows researchers to test the relevance of any cost model for real spatial data sets. By calculating back from microscopy data, they can for the first time check different classes of the model for plausibility. Physicist Carl Modes, who oversaw the study says: “This model approach is not limited to the liver, but may be applied to and tested on any entangled system such as pancreas, kidney, lymphatic system or bone marrow. Although there are still limitations on the estimation of either of the stimuli (blockage or enhancement), improving its applicability and quality may help to properly classify and identify crucial stimuli in arbitrary biological flow networks. Our ultimate dream would be the identification of pathological deformities in the vessel systems, and making it a diagnostic tool e.g. for liver cirrhosis, detecting polycystic diseases (e.g. in kidney), or tumor growth.”
Felix Kramer and Carl D. Modes: "How to pare a pair: Topology control and pruning in intertwined complex networks", Phys. Rev. Research 2, 043171, published 2 November 2020
DOI: 10.1103/PhysRevResearch.2.043171