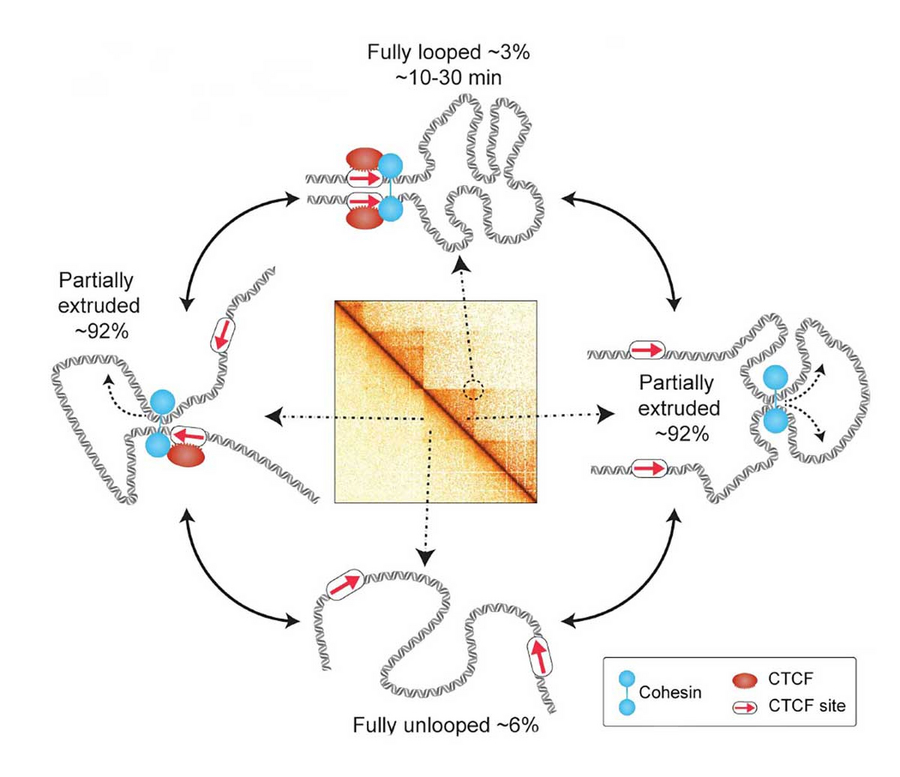
Super-resolution live-cell imaging of CTCF/Cohesin-mediated chromatin loops reveals these loops to be both rare and dynamic. © Gabriele et al., Science 10.1126/science.abn6583 (2022).
In human chromosomes, DNA is coated by proteins to form an exceedingly long beaded string. This “string” is folded into numerous loops, which are believed to help cells control gene expression and facilitate DNA repair, among other functions. A new study from the Massachusetts Institute of Technology (MIT) in collaboration with the Max Planck Institute of Molecular Cell Biology and Genetics (MPI-CBG) in Dresden, Germany and the Center for Systems Biology Dresden (CSBD) suggests that these loops are very dynamic and shorter-lived than previously thought. In the new study, the researchers were able to monitor the movement of one stretch of the genome in a living cell for about two hours. They saw that this stretch was fully looped for only 3 to 6 percent of the time, with the loop lasting for only about 10 to 30 minutes. The findings suggest that scientists’ current understanding of how loops influence gene expression may need to be revised, the researchers say.
“Many models in the field have been these pictures of static loops regulating these processes. What our new paper shows is that this picture is not really correct,” says Anders Sejr Hansen, the Underwood-Prescott Career Development Assistant Professor of Biological Engineering at MIT. “We suggest that the functional state of these domains is much more dynamic.” Hansen is one of the senior authors of the new study, along with Leonid Mirny, a professor in MIT’s Institute for Medical Engineering and Science and the Department of Physics, and Christoph Zechner, a group leader at the MPI-CBG and the CSBD. MIT postdoc Michele Gabriele, recent Harvard University PhD recipient Hugo Brandão, and MIT graduate student Simon Grosse-Holz are the lead authors of the paper, which appears today in Science.
Out of the loop
Using computer simulations and experimental data, scientists including Mirny’s group at MIT have shown that loops in the genome are formed by a process called extrusion, in which a molecular motor promotes the growth of progressively larger loops. The motor stops each time it encounters a “stop sign” on DNA. The motor that extrudes such loops is a protein complex called cohesin, while the DNA-bound protein CTCF serves as the stop sign. These cohesin-mediated loops between CTCF sites were seen in previous experiments. However, those experiments only offered a snapshot of a moment in time, with no information on how the loops change over time. In their new study, the researchers developed techniques that allowed them to fluorescently label CTCF DNA sites so they could image the DNA loops over several hours. They also created a new computational method that can infer the looping events from the imaging data. “This method was crucial for us to distinguish signal from noise in our experimental data and quantify looping,” Zechner says. “We believe that such approaches will become increasingly important for biology as we continue to push the limits of detection with experiments.”
The researchers used their method to image a stretch of the genome in mouse embryonic stem cells. “If we put our data in the context of one cell division cycle, which lasts about 12 hours, the fully formed loop only actually exists for about 20 to 45 minutes, or about 3 to 6 percent of the time,” Grosse-Holz says.
“If the loop is only present for such a tiny period of the cell cycle and very short-lived, we shouldn't think of this fully looped state as being the primary regulator of gene expression,” Hansen says. “We think we need new models for how the 3D structure of the genome regulates gene expression, DNA repair, and other functional downstream processes.” While fully formed loops were rare, the researchers found that partially extruded loops were present about 92 percent of the time. These smaller loops have been difficult to observe with the previous methods of detecting loops in the genome. “In this study, by integrating our experimental data with polymer simulations, we have now been able to quantify the relative extents of the unlooped, partially extruded, and fully looped states,” Brandão says.
“Since these interactions are very short, but very frequent, the previous methodologies were not able to fully capture their dynamics,” Gabriele adds. “With our new technique, we can start to resolve transitions between fully looped and unlooped states.”
The researchers hypothesize that these partial loops may play more important roles in gene regulation than fully formed loops. Strands of DNA run along each other as loops begin to form and then fall apart, and these interactions may help regulatory elements such as enhancers and gene promoters find each other. “More than 90 percent of the time, there are some transient loops, and presumably what's important is having those loops that are being perpetually extruded,” Mirny says. “The process of extrusion itself may be more important than the fully looped state that only occurs for a short period of time.”
More loops to study
Since most of the other loops in the genome are weaker than the one the researchers studied in this paper, they suspect that many other loops will also prove to be highly transient. They now plan to use their new technique study some of those other loops, in a variety of cell types. “There are about 10,000 of these loops, and we've looked at one,” Hansen says. “We have a lot of indirect evidence to suggest that the results would be generalizable, but we haven’t demonstrated that. Using the technology platform we’ve set up, which combines new experimental and computational methods, we can begin to approach other loops in the genome.” The researchers also plan to investigate the role of specific loops in disease. Many diseases, including a neurodevelopmental disorder called FOXG1 syndrome, could be linked to faulty loop dynamics. The researchers are now studying how both the normal and mutated form of the FOXG1 gene, as well as the cancer-causing gene MYC, are affected by genome loop formation.
The research was funded by the National Institutes of Health, the National Science Foundation, the Mathers Foundation, a Pew-Stewart Cancer Research Scholar grant, the Chaires d'excellence Internationale Blaise Pascal, an American-Italian Cancer Foundation research scholarship, and the Max Planck Institute for Molecular Cell Biology and Genetics.
Video demonstrating chromosome organization by loop extrusion with barrier elements by the Mirny lab: https://www.youtube.com/watch?v=8FW6gOx5lPI
Reprinted with permission of MIT News
Original MIT News Release
Michele Gabriele, Hugo B. Brandão, Simon Grosse-Holz, Asmita Jha, Gina M. Dailey, Claudia Cattoglio, Tsung-Han S. Hsieh, Leonid Mirny, Christoph Zechner, Anders S. Hansen: Dynamics of CTCF and cohesin mediated chromatin looping revealed by live-cell imaging. Science, 14. April 2022, doi: 10.1126/science.abn6583
Anders Sejr Hansen
+1 617-253-6086
ashansen(at)mit.edu
Christoph Zechner
+49 (0) 351 210-2735
zechner(at)mpi-cbg.de