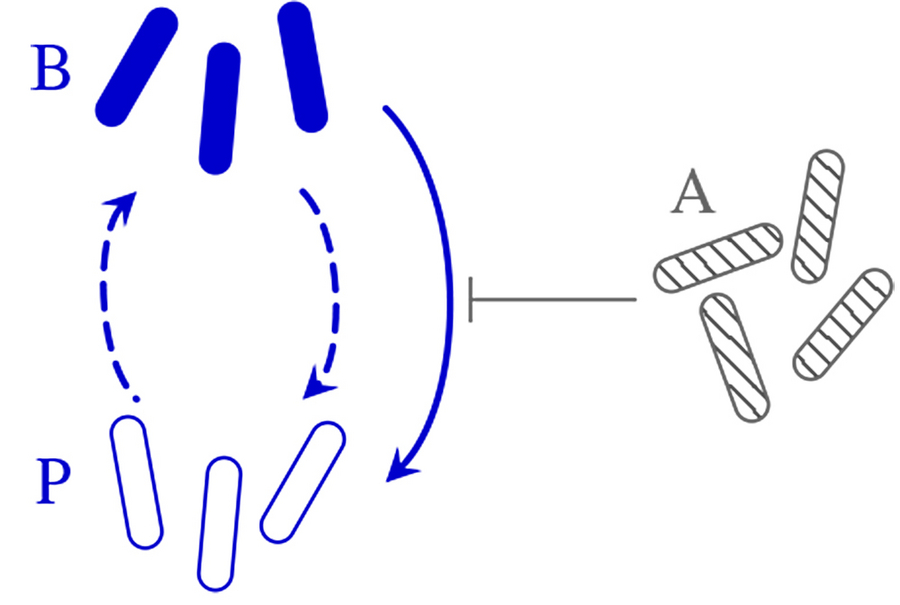
Minimal setup for responsive switching: a bacterial species has two subgroups B and P between which it switches stochastically. Additionally, the B subgroup of "normal" bacteria switches to the slowly dividing, resilient "persister" subgroup P in the presence of the competitor species A. © Haas et al., Phys. Rev. Res., 2022
The different species of microbes in complex ecological communities such as the human microbiome often have different subgroups called phenotypes, between which microbes can switch stochastically or in response to environmental cues, such as toxins released by competitors or antibiotics. Pierre Haas, research group leader at the Max Planck Institute for the Physics of Complex Systems and the Max Planck Institute of Molecular Cell Biology and Genetics, who is also affiliated with the Center for Systems Biology Dresden, analysed the ecological implications of such responsive switching with collaborators at the University of Cambridge. The researchers combined a statistical analysis of many-species systems, a numerical study of a minimal two-species model, and exact results for still simpler mathematical models of this responsive switching. While responsive switching to a rare phenotype is generally destabilising, they could show that responsive switching to a rare "attack" phenotype is stabilising on average. A similar "attack" subgroup of a bacterial species was recently observed experimentally, which underlines the importance of responsive switching for ecological stability.
Pierre A. Haas, Maria A. Gutierrez, Nuno M. Oliveira, and Raymond E. Goldstein, "Stabilization of microbial communities by responsive phenotypic switching", Phys. Rev. Research 4, 033224., doi.org/10.1103/PhysRevResearch.4.033224