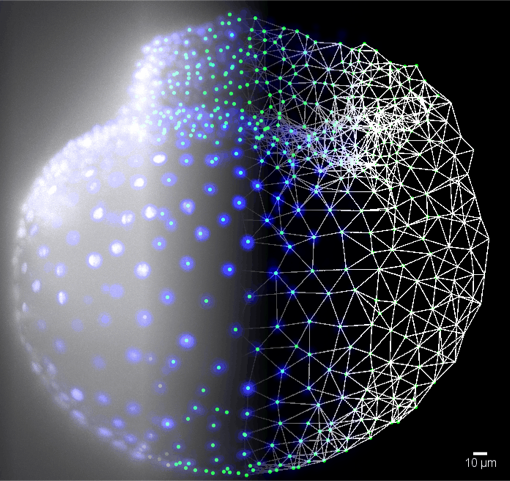
Schematic view of processing imaging data with CLIJ: raw light sheet microscopy data of red flour beetle embryo Tribolium castaneum is processed by background subtraction (blue), spot detection (green), to finally generate surface reconstruction mesh data (white, right) describing the embryo's shape. © Robert Haase, CSBD/MPI-CBG, Imaging credits: Daniela Vorkel
Answering fundamental biological questions involves state-of-the-art imaging technologies that generate huge amounts of image data. For example, quantitative analysis of long time lapse movies of developing fruit fly embryos acquired with a light sheet microscope, can take days of processing time. It has therefore become more and more apparent, that there is a strong need for accelerating image processing in a general way.
Graphics processing units (GPUs) allow image processing at unprecedented speed. But utilizing GPUs for scientific purposes is not trivial. The potential benefits of GPU-acceleration are well known and exploited on a project-by-project basis all over the world. In ImageJ/Fiji, a widespread software application among biomedical researchers, a generic GPU-accelerated image processing toolkit has thus far been missing.
Computer scientists from the Gene Myers group at the The Center for Systems Biology Dresden (CSBD) and the MPI-CBG, together with colleagues from the Chan Zuckerberg Biohub in San Francisco, have now created such a toolkit, called CLIJ for both ImageJ and Fiji. CLIJ can reduce the processing time of typical image processing workflows by orders of magnitude. What took hours before can now be done in minutes. In their recent publication in Nature Methods, the researchers quantify in detail the acceleration potential of GPU-based image processing.
The novel software is freely available, completely open-source, and particularly user-friendly. It was published together with an extraordinarily detailed user documentation, example codes, and explanations for various programming languages. It thus lowers the entry-level for scientists interested in GPU-acceleration. Furthermore, CLIJ runs on graphics cards from the major vendors Intel, AMD and NVidia and under the major operating systems Windows, MacOS and Linux.
Robert Haase, first author of the paper, says, “We were able to show that some CLIJ-operations ran up to about 200 times faster on the GPU than their counter part ImageJ-operations. A speedup factor of 200 means, what took three months before, can now be done in a single day… before lunch. We developed CLIJ completely in the open together with the community. One of our major motivations was to enable scientists to take advantage of GPU-acceleration without the need to involve high-performance-computing experts.”
The supervisor of the work, Gene Myers, adds, “We can already see what impact CLIJ has in the scientific community. It can enable projects, which couldn’t be done yet because they simply took too much time. According to early adopters, CLIJ is a game changer.”

Verarbeitung von Bilddaten mit CLIJ © Robert Haase, CSBD/MPI-CBG, Bildnachweis: Daniela Vorkel
Robert Haase, Loic Alain Royer, Peter Steinbach, Deborah Schmidt, Alexandr Dibrov, Uwe Schmidt, Martin Weigert, Nicola Maghelli, Pavel Tomancak, Florian Jug, Eugene W Myers. "CLIJ: GPU-accelerated image processing for everyone." Nature Methods, 18 November, 2019, DOI: 10.1038/s41592-019-0650-1