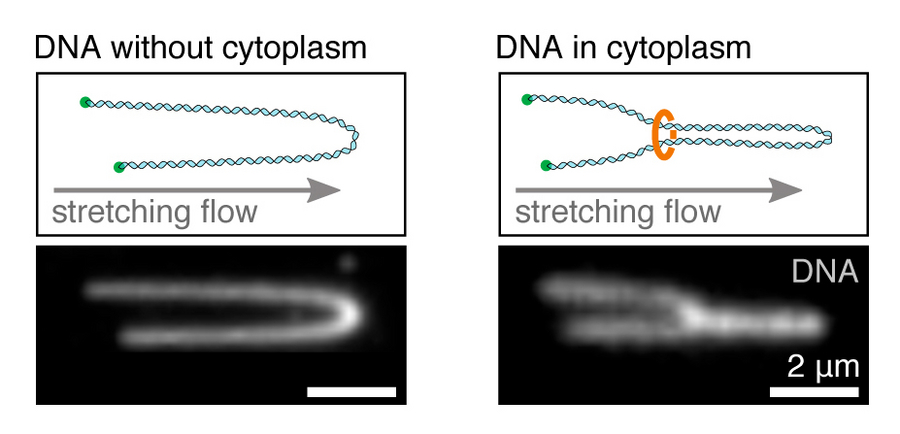
Stretching of DNA loops. Left: stretching a single, immobilised DNA molecule in buffer. Right: Stretched DNA molecule showing a loop, mediated by ring-shaped protein complexes in the cytoplasm. Copyright: Golfier et al. / MPI-CBG
With more than 3 billion base pairs, the human genome in each cell nucleus stretches to a physical length of two meters. How cells organize those vast amounts of genomic material inside a micrometer-sized nucleus without making knots and tangles has been a mystery. Extrusion of DNA loops through ring shaped protein complexes has been proposed as a mechanism that spatially organizes the genome in all living organisms. However, a direct proof for the existence of those loops in a living context was missing. Researchers at the Max Planck Institute of Molecular Cell Biology and Genetics (MPI-CBG) and the MPI for the Physics of Complex Systems, both in Dresden, have now, for the first time, observed DNA loop extrusion in a living context. The study was published in the journal eLife.
The combined length of all DNA molecules inside the cells of a human body would equal the diameter of our solar system, about 10 billion kilometres. In order for organisms to develop correctly and stay healthy throughout life, the genomic material inside the cell nucleus has to be spatially organized without knots and tangles. This task is equivalent to trying to fit about 100 kilometres of thin hair knot-free in a football. How cells achieve this complicated organizational task, has been a mystery. Over the last decade, a model was proposed, that seems to solve this riddle by assuming that ring-shaped protein complexes can bind to DNA and actively extrude DNA into a loop. Those loops prevent knots and tangles and also help to regulate genes, which is important for the development and health of an organism. Computer simulations of this ‘DNA loop extrusion’ model were able to recapitulate many features seen in experiments. Additionally, the formation of DNA loops could recently be observed in experiments employing purified ring-shaped protein complexes onto single DNA molecules. However, a direct proof that loops spatially organise DNA natively in a living environment was missing.
Scientists from the group of Jan Brugués at the MPI-CBG and the MPI-PKS now developed a method to visualize DNA loop extrusion for the first time in a living context. They were able to reduce the complexity of the cell nucleus by taking small pieces of DNA and stretching them out in the cytoplasm (the living liquid content of cells), extracted from eggs of the African clawed frog (Xenopus laevis). Stefan Golfier, the first author of the study, explains: “With the help of Total Internal Reflection Fluorescence (TIRF) microscopy, a great method for observing single molecules in real-time, we were able to show that cohesin and condensin, the proposed ring-shaped protein complexes that are present in the native cytoplasm, spatially organise DNA into loops. This process takes place both during cell growth (the interphase) and cell division (metaphase). Intriguingly, the mechanism and responsible motors that establish DNA loops differ during these cell cycle phases. In interphase, when chromatin is openly organized into compartments that regulate gene expression, DNA is pulled into a loop from both left and right through the cohesin-ring. However, during metaphase, where chromosomes undergo large-scale compaction, DNA loops are established by condensin, which pulls DNA through the ring from only one side to make a loop.”
Jan Brugués, who oversaw the study, concludes: “Our work provides the first direct evidence and biophysical characterization of native DNA loop formation as a mechanism to spatially organize DNA in the cytoplasm of a higher vertebrate. As the responsible protein complexes are highly conserved across species, our findings suggest, that DNA loop extrusion might be a general mechanism to organize genomes in all life forms on earth.” He continues: “Understanding the underlying mechanisms of how the genome is folded will contribute to a better understanding and treatment of diseases that are associated with distortions in the architecture of the genome, such as developmental defects and cancer.” This assay could further help expanding our understanding of the genome function, as it allows to build the complexity of the genome from the bottom up.
-----------
Jan Brugués is also affiliated with the Center for Systems Biology Dresden (CSBD) and the Cluster of Excellence “Physics of Life” (PoL) at the TU Dresden. The CSBD is a cooperation between the MPI-CBG, the MPI-PKS and the TU Dresden. In the interdisciplinary center, physicists, computer scientists, mathematicians, and biologists work together to understand how cells coordinate their behaviour to form tissues and organs of a given form or function. PoL is focusing on fundamental organizational principles of living matter through a close interaction of experiment and theory combined with computational modeling, simulation and interactive microscopy.
Stefan Golfier, Thomas Quail, Hiroshi Kimura, Jan Brugués “Cohesin and condensin extrude DNA loops in a cell-cycle dependent manner” eLife, 12. May 2020. Doi: 10.7554/eLife.53885