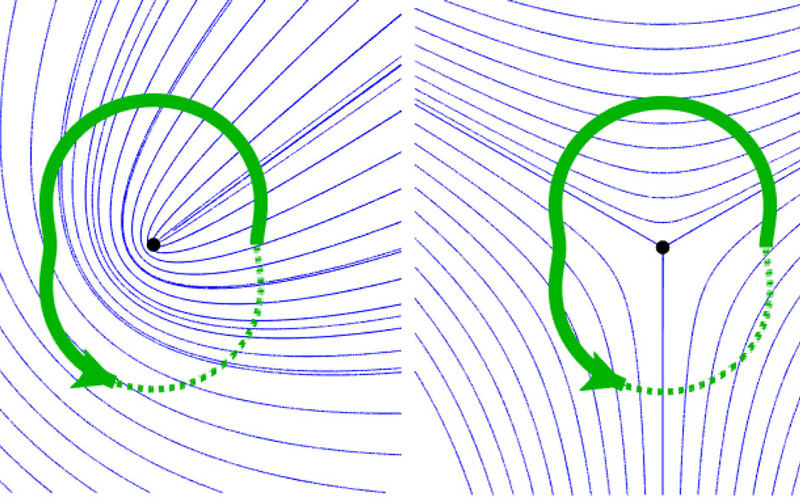
Orientation field (blue lines) with topological defect (black dot). Identification of the defect by an encircling path (green), along which all possible orientations are encountered © Karl Hoffmann, CSBD / MPI-CBG
With the constant development of new state-of-the-art microscopy techniques, the quantitative analysis of image data is becoming increasingly important for researchers in the life sciences. For the description of directed objects and processes like cell elongation or the structure of the cytoskeleton, physicists use orientation fields. Such fields describe for example the orientation of cytoskeletal fibers. As motor proteins “walk” along the cytoskeleton and generate force, the orientation field informs researchers on the forces acting in the biological tissue they observe under their microscopes. Topological defects are special points in orientation fields, where the field has a kind of “jump”. One can say, the orientation field rotates around a topological defect such that it attains all possible directions in the vicinity of the defect. The order in which the rotation happens and the number of full rotations (called topological charge) are used to identify topological defects in data.
Topological defects are deeply linked to shape and curvature. They are used to characterize cell stress and predict the locations where a flat sheet of cells will buckle or extrude a cell. So far, it wasn’t known, to what extent topological defects identified in noisy data are actually describing the reality or just mirror the amount of noise in the data. In other words, the question was how much can you trust topological defects identified in data? Researchers in the group of Ivo Sbalzarini, group leader at the MPI-CBG and the Center for Systems Biology Dresden (CSBD), and professor of Scientific Computing for Systems Biology at the TU Dresden, have now developed a new mathematical answer to this question and published their findings in Physical Review E.
Karl Hoffmann, a final-year Predoc in Ivo’s group and first author of the paper explains, “When studying noisy Drosophila (fruit fly) microscopy images, I realized that we have to refine our question first. We then focused on quantifying by how much the orientation field can change without an alteration of the topological defects observed. In other words, at which point does the topological defect switch when the image contains noise or due to the temporal evolution of the tissue?” The mathematician continues, “Then, I looked at the orientation differences between any two pixels in my image data, and how they build up to indicate a topological defect (or none). With our method, scientists can now quantify the reliability of detected defects, filter spurious ones from the data, and predict from a static image the possible dynamics of topological defects like motion or two of them merging and disappearing.
Ivo Sbalzarini adds, “Topological defects are increasingly used by several groups at the CSBD and worldwide as a coarse-grained observable of how biological tissues grow to their final shape, yet so far nothing was known about their robustness. Our result will benefit the analysis of topological defects in microscopy image and simulation data alike. It allows to capture the influence of noise, which is of special importance to biophysical models, and can help us understand fundamental biological questions, for example how robustly the shape and form of a tissue is predetermined by the genes, or how organisms can develop robustly in the face of unpredictable environmental influences and variability between individuals.”
Karl B. Hoffmann and Ivo F. Sbalzarini: "Robustness of topological defects in discrete domains", Phys. Rev. E 103, 012602, published 5 January 2021, DOI:https://doi.org/10.1103/PhysRevE.103.012602