Stephan Saalfeld
Tiled serial section transmission electron microscopy (ssTEM) is increasingly used to describe high-resolution anatomy of large biological specimens. In particular, it is indispensable for analysis of the neural tissue at the level of synaptic connectivity. Our collaborator, Albert Cardona, collects massive ssTEM dataset covering the ENTIRE first instar larval nervous system consisting of several hundred thousands image tiles.
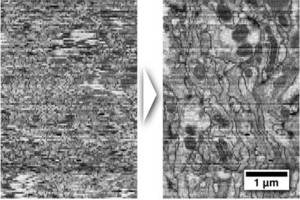
Registration of serial section TEM image mosaics has to preserve the continuity of the 3d-structure of the specimen while compensating the physical distortions applied to the tissue during sectioning and imaging, staining artifacts, missing sections and the fact that certain structures may appear dissimilar in consecutive sections. We first developed a fully automatic, as-rigid-as-possible registration approach for large tiled serial section microscopy stacks that uses Scale Invariant Feature Transform (SIFT) to identify corresponding landmarks within and across sections and globally optimize the pose of each single tile for minimal overall displacement of these landmark correspondences (Saalfeld et al. 2010). Subsequently, we have developed an elastic alignment approach that takes the SIFT based alignment as starting point and attempts to remove non-linear distortion introduced during section preparation while preserving the true shape of the structures within the tissue (Saalfeld et al. 2012). We demonstrate the performance of the elastic alignment on two outstanding ssTEM section series:one segment of Drosophila first instar larval ventral nerve cord consisting of nearly 33,000 images and the entire 558 cell C. elegans embryo (data courtesy of Rick Fetter Janelia Farm). See the spectacular movies visualizing the registration results.
We also developed CATMAID, a web-based tool modeled after GoogleMaps, for collaborative annotation of spatially large image datasets (Saalfeld et al. 2009). This tools is actively used by Open Connectome to show some of the largest TEM datasets in existence. The tool is further developed for collaborative neuronal circuits reconstruction by the lab of Albert Cardona. We plan to extend it for viewing, sharing and collaborative annotation of multi-dimensional light microscopy datasets.
collaborators: Albert Cardona (ETH Zurich/Janelia Farm), Volker Hartenstein (UCLA)
funding: MPI-CBG
Saalfeld S., Cardona A., Hartenstein V., Tomancak, P. (2010) As-rigid-as-possible mosaicking and serial section registration of large ssTEM datasets Bioinformatics, 26(12), 57-63
Cardona A., Saalfeld S., Preibisch S., Schmid B., Cheng A., Pulokas J., Tomancak P., Hartenstein V. (2010) An integrated micro- and macroarchitectural analysis of the Drosophila brain by computer-assisted serial section electron microscopy. PLoS Biology 8(10): e1000502
Saalfeld S., Fetter R., Cardona A., Tomancak P. (2012) Elastic Volume Reconstruction from Series of Ultra-thin Microscopy Sections Nature Methods 9(7), 717-720
Saalfeld S., Cardona A., Hartenstein V., Tomancak P. (2009) CATMAID: Collaborative Annotation Toolkit for Massive Amounts of Image Data. Bioinformatics Aug 1;25(15):1984-6