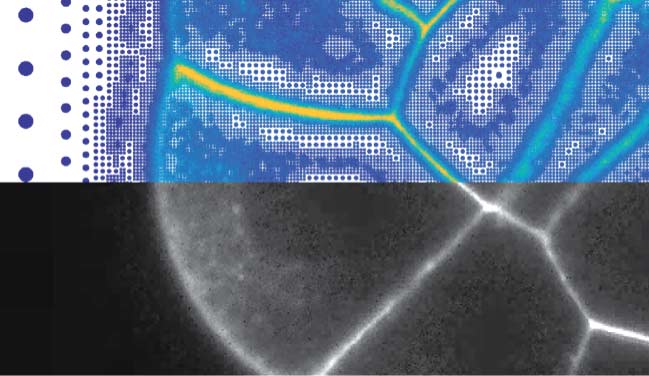
Representing fluorescence images with particles instead of pixels © Sbalzarini Group, MPI-CBG, CSBD
Microscopy is the main tool for investigation in biomedical research. In the past years, modern microscopes have developed to semi-automatic computer-driven machines that can record 3D images at very high resolution over the course of many hours or even days. This technological advance opens new opportunities for biomedical research and has been at the root of many recent break-through discoveries. However, the cutting-edge microscopes generate more data than can reasonably be handled. With gigabytes per second, or terabytes per day being created, storing and processing the images becomes a severe challenge and is often the limiting factor in fully using the capabilities of modern microscopes.
Researchers around Ivo Sbalzarini, Research Group Leader at the MPI-CBG, and the Center for Systems Biology Dresden (CSBD), have now found an innovative way to address this problem. Sbalzarini’s team argued that the data problem is caused by storing images as grids of pixels. To solve this, the computer scientists developed a method that replaces pixels with particles, that are positioned according to the image content. The novel approach, called Adaptive Particle Representation (APR), places particles only where there is content in the image, and uses particles of different sizes to reflect the local degree of detail. This way, the storage and processing costs of images can be reduced by several hundred-fold. For example, the APR of a 30 gigabytes image that required five hours to be processed is just about 50 megabytes, so that the image can be processed in 30 seconds. The new method, recently published in Nature Communications, includes a software package, that is freely available and designed to be used and adopted by anyone.
Bevan L. Cheeseman, first author of the publication, says, “Our approach is of interest to anyone acquiring, storing, or processing fluorescence microscopy images. It eliminates “data waste” and utilizes microscopes and computer power more efficiently by orders of magnitude.”
Sbalzarini, who is also a Professor of Computer Science at TU Dresden, adds, “The APR is not only good for the speed at which images can be processed or visualized, and for reducing the money spent on hard disks. It also has a positive effect on the climate, as several hundred-fold less electricity is consumed to perform a given image-analysis task, resulting in a lower carbon footprint of image-based biology research. We are very happy that the APR combines advancing biomedical imaging with significant energy savings.”
B. L. Cheeseman, U. Günther, K. Gonciarz, M. Susik, and I. F. Sbalzarini. Adaptive particle representation of fluorescence microscopy images. Nature Communications, 9:5160, 2018. DOI: 10.1038/s41467-018-0739