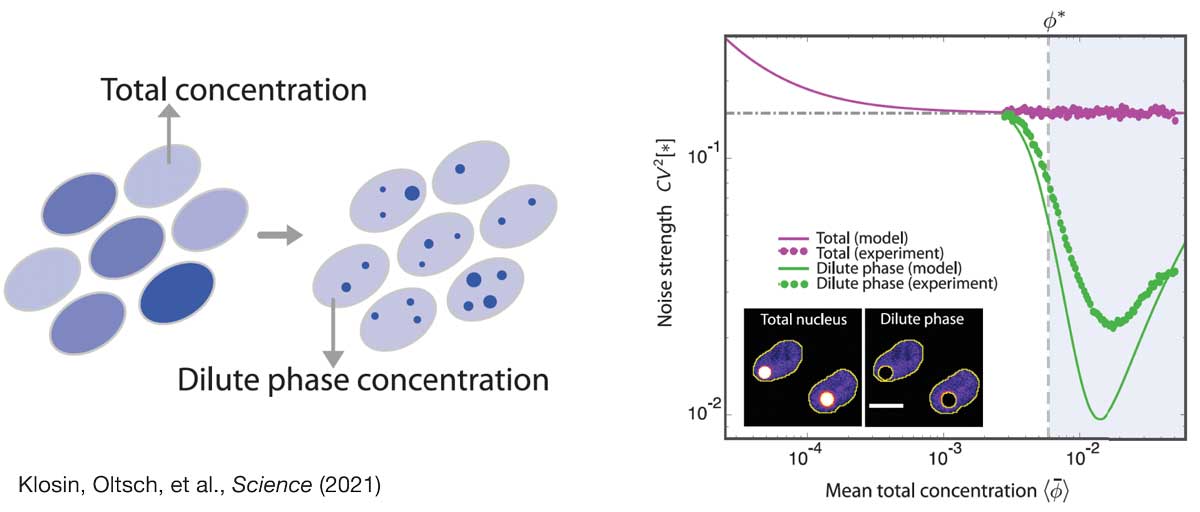
In this project, we study the role of biomolecular condensates to control intracellular noise. We have recently provided a first proof-of-principle of this idea, showing that protein concentration noise can be strongly reduced when the protein partitions into condensates. Based on these findings we are now exploring the generality of this concept in the context of cellular information processing and feedback control. To this end, we merge statistical physics with control theory to understand the statistical constraints of chemical pathways in condensed, non-equilibrium environments. We complement our theoretical work with experiment in close collaboration with the Hyman lab.
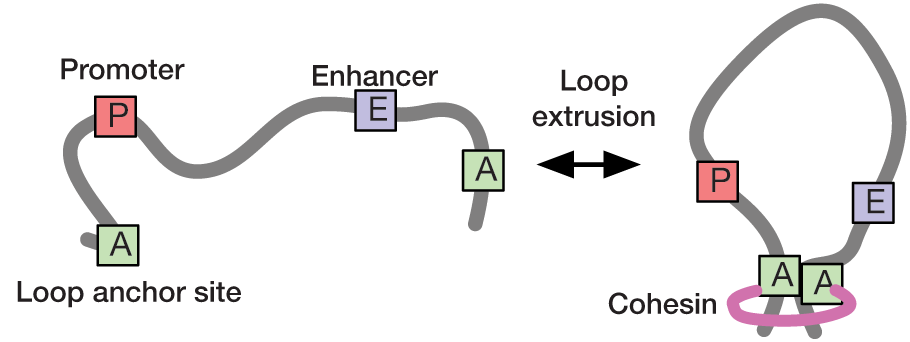
Loop extrusion has been proposed as a mechanism to compartmentalize chromatin into topologically associating domains (TADs), thereby facilitating interactions between promoters and enhancers. In collaboration with the Hansen and Mirny labs at MIT, we use statistical modelling and super-resolution live-cell imaging to understand the dynamics of chromatin looping and its role in transcription regulation. We have recently developed a rigorous statistical method to infer loop contact frequencies and lifetimes from noisy time-series data. Our long-term goal is to use these approaches to establish a quantitative link between the dynamics of chromatin looping and transcription.