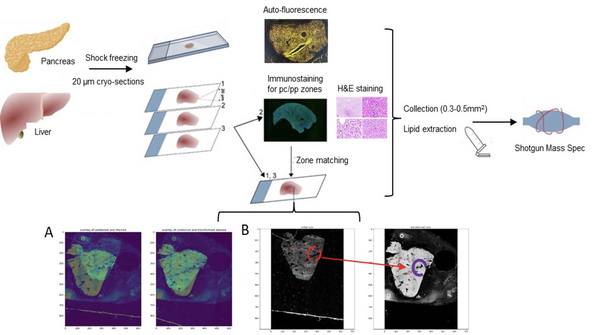
Quantitative omics characterization of spatially defined areas with altered tissue morphology could reveal specific lipidomics and proteomics markers of progressing hepatocellular carcinoma (HCC), colon cancer (CRC), metabolic diseases (NASH) or diabetes [1]. To this end, we combined quantitative shotgun lipidomics and LC-MS/MS proteomics with laser-capture microdissection (LCM) in a single multi-omics approach to assess the spatial heterogeneity of the lipidome and proteome in the pathological and surrounding tissues [2].
Histological staining of cryo-sections may bias their lipid composition. Therefore, if needed we stain and image a separate cryo-section, overlay the image onto the unstained section and used it for LCM isolation of visualized features. Images matching is supported by Jupiter Notebook based software (A-B Figure below, [3]).
LCM-omics powered by the in-house developed software is used for the systematic and consistent characterization of hepatic, intestinal and pancreatic biopsies.
1.Ucal, Y. et al., Bioch. Biophys. Acta, 2017, 1865; 795-816
2.Shevchenko, A. et al., Nat. Rev. Mol. Cell Biol, 2010, 11; 593-598
3.Knittelfelder, O. et al., Anal Chem, 2018, 90; 9868-9878 [modified]
Our main collaborators for LCM-lipidome studies : Dr. Michele Solimena’s lab (Paul Langerhans Institute, Dresden); Dr. Sebastian Zeissig’s lab (Center for Regenerative Therapies, Dresden); Dr. Jochen Hampe’s lab (Universitätsklinikum Carl Gustav Carus, Dresden) and LiSym consortium; Dr. Marino Zerial’s lab (MPI-CBG).
Olga Vvedenskaya, Tim Daniel Rose, Oskar Knittelfelder, Alessandra Palladini, Judith Andrea Heidrun Wodke, Kai Schuhmann, Jacobo Miranda Ackerman, Yuting Wang, Canan Has, Mario Brosch, Veera Raghavan Thangapandi, Stephan Buch, Thomas Züllig, Jürgen Hartler, Harald C Köfeler, Christoph Röcken, Ünal Coskun, Edda Klipp, Witigo von Schoenfels , Justus Gross, Clemens Schafmayer, Jochen Hampe, Josch Konstantin Pauling, Andrej Shevchenko. Nonalcoholic fatty liver disease stratification by liver lipidomics. J Lipid Res., doi:10.1016/j.jlr.2021.100104. (2021). DOI
Veera Raghavan Thangapandi, Oskar Knittelfelder, Mario Brosch, Eleonora Patsenker, Olga Vvedenskaya, Stephan Buch, Sebastian Hinz, Alexander Hendricks, Marina Nati, Alexander Herrmann, Devavrat Ravindra Rekhade, Thomas Berg, Madlen Matz-Soja, Klaus Huse, Edda Klipp, Josch K Pauling, Judith Ah Wodke, Jacobo Miranda Ackerman, Malte von Bonin, Elmar Aigner, Christian Datz, Witigo von Schönfels, Sophie Nehring, Sebastian Zeissig, Christoph Röcken, Andreas Dahl, Trian Chavakis, Felix Stickel, Andrej Shevchenko, Clemens Schafmayer, Jochen Hampe, Pallavi Subramanian. Loss of hepatic Mboat7 leads to liver fibrosis. Gut, Art. No. doi: 10.1136/gutjnl-2020-320853 (2020) DOI
Yuting Wang, Sebastian Hinz, Ortrud Uckermann, Pia Hönscheid, Witigo von Schönfels, Greta Burmeister, Alexander Hendricks, Jacobo Miranda Ackerman, Gustavo Baretton, Jochen Hampe, Mario Brosch, Clemens Schafmayer, Andrej Shevchenko, Sebastian Zeissig. Shotgun lipidomics-based characterization of the landscape of lipid metabolism in colorectal cancer. Biochim Biophys Acta Mol Cell Biol Lipids, 1865(3) Art. No. 158579 (2020) DOI
Olga Vvedenskaya, Yuting Wang, Jacobo Miranda Ackerman, Oskar Knittelfelder, Andrej Shevchenko. Analytical challenges in human plasma lipidomics: A winding path towards the truth. Trends Analyt Chem, Art. No. 115277 (2018) DOI
Susanne Sales, Juergen Graessler, Sara Ciucci, Rania Al-Atrib, Terhi Vihervaara, Kai Schuhmann, Dimple Kauhanen, Marko Sysi-Aho, Stefan Bornstein, Marc Bickle, Carlo Vittorio Cannistraci, Kim Ekroos, Andrej Shevchenko. Gender, Contraceptives and Individual Metabolic Predisposition Shape a Healthy Plasma Lipidome. Sci Rep, 6 Art. No. 27710 (2016) PDF DOI
Susanne Sales, Oskar Knittelfelder, Andrej Shevchenko. Lipidomics of Human Blood Plasma by High-Resolution Shotgun Mass Spectrometry. Methods Mol Biol, 1619 203-212 (2017) DOI
Santosh Phuyal, Mayes Kasem, Oskar Knittelfelder, Animesh Sharma, Davi de Miranda Fonseca, Vaineta Vebraite, Sergey Shaposhnikov, Geir Slupphaug, Vidar Skaug, Shanbeh Zienolddiny. Characterization of the proteome and lipidome profiles of human lung cells after low dose and chronic exposure to multiwalled carbon nanotubes.
Nanotoxicology, 12(2) 138-152 (2018) DOI
Oskar Knittelfelder, Sofia Traikov, Olga Vvedenskaya, Andrea Schuhmann, Sandra Segeletz, Anna Shevchenko, Andrej Shevchenko. Shotgun Lipidomics Combined with Laser Capture Microdissection: A Tool To Analyze Histological Zones in Cryosections of Tissues. Anal Chem, 90(16) 9868-9878 (2018) DOI
Hample lab Coskun lab Klipp lab Pauling lab and LiSyM
Solimena Lab Matz-Soja lab Zeissig lab Cramer lab
